The biology of SARS-CoV-2, the virus behind the COVID-19 pandemic, remains partially elusive. Understanding viral mechanisms is a key factor in developing effective treatment strategies against the outbreak.
Now, Keesiang Lim and Richard Wong from Kanazawa University and colleagues have shown how the virus is equipped to enter human cells in real-time.
SARS-CoV-2 is enveloped by spike proteins, which form a crown-like layer on its surface. The immune system detects these spike proteins and prepares to neutralize the virus. Spike proteins also play a role in mediating entry of SARS-CoV-2 into cells.
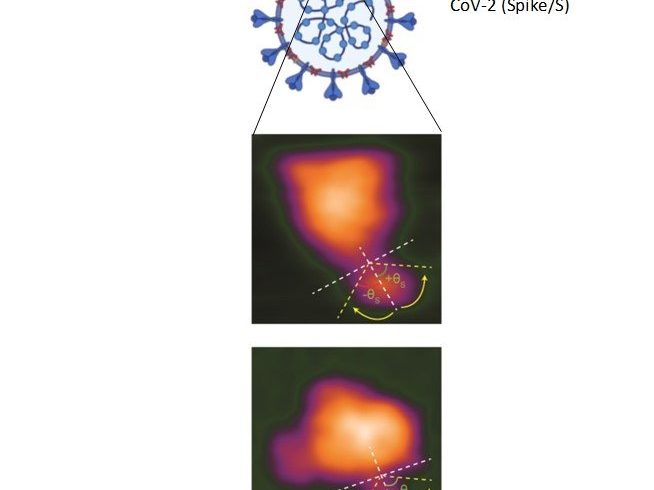
To date, scientists have been able to take high-resolution albeit stationary images of spike proteins. Importantly, Richard Wong’s team and Kanazawa University used an advanced form of microscopy to capture dynamic changes in spike proteins when bound to cells.
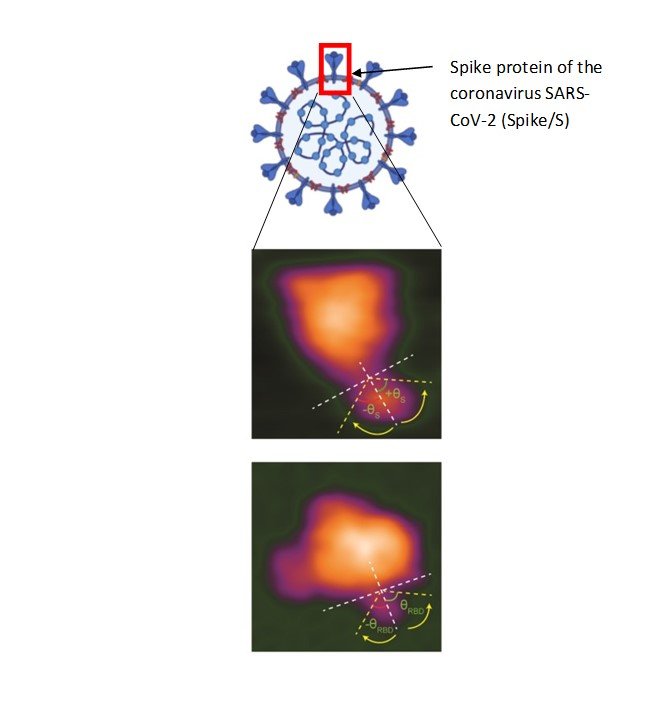
Spike proteins comprise two major components—a globular head (that has a host recognition domain) attached to a stalk (that is capable of fusing with cells and facilitating entry). The researchers used high-speed atomic force microscopy (HS-AFM) to understand this structure deeper using just single molecules of spike proteins.
“We previously showed that the real-time observation of structural dynamic of influenza A hemagglutinin during viral entry,” says lead author of the study Dr. Keesiang Lim.
In the newly-published study, they found that the stalk showed a very flexible nature with an ability to extend or retract whereas the head could change conformation resulting in disappearance of the host recognition domain.
Spike proteins typically latch onto cells that exhibit a molecule called ACE2 on their surface. Thus, the interactions of spike proteins with ACE2 were then visualized by HS‑AFM. It was found that spike proteins docked onto ACE2 with the host recognition domain exposed. What’s more, their elastic nature enabled a far smoother interaction.
Small extracellular vesicles (sEV) are sacs released by cells which are composed of the similar chemical constituents as the cell membrane. The dynamics of spike proteins on sEVs was studied next. Since the spike protein stalk facilitates binding and fusion with membranes, interactions of only the stalk with sEVs were analyzed first.
Indeed, a disruption of sEV membranes was seen indicating the stalk could fuse with cell membranes easily. However, when the dynamics of the entire spike protein were assessed, stable binding was only observed with sEVs released from ACE2-containing cells. ACE2 was thus a key factor in mediating viral entry.
HS-AFM proved to be a very useful tool in understanding the entry mechanics of SARS‑CoV-2 in detail. “Altogether, our study provides a platform that is suitable for real-time visualization of various entry inhibitors, neutralizing antibodies, and sEV-based decoy in blocking viral entry,” explains Dr. Richard Wong, senior author of the study. Blocking the connections between spike proteins and ACE2 or inhibiting the membrane disruption caused by the spike protein stalk could be potential strategies preventing SARS‑CoV-2 from hijacking the body.