Ribosomes are the complexes of ribonucleoproteins at the heart of protein synthesis in cells. However in the absence of conclusive evidence, how these complexes operate has been open to debate.
Now Hirotatsu Imai and Noriyuki Kodera at Kanazawa University, alongside Toshio Uchiumi at Niigata University in Japan, show visualizations of the structural dynamics and factor pooling that take place at ribosome stalk proteins as they build new proteins.
Ribosomes were first discovered in the 1950s and their broad function has been widely understood for some time – they read messenger RNA sequences and from that generate sequences of correctly ordered amino acids into new proteins.
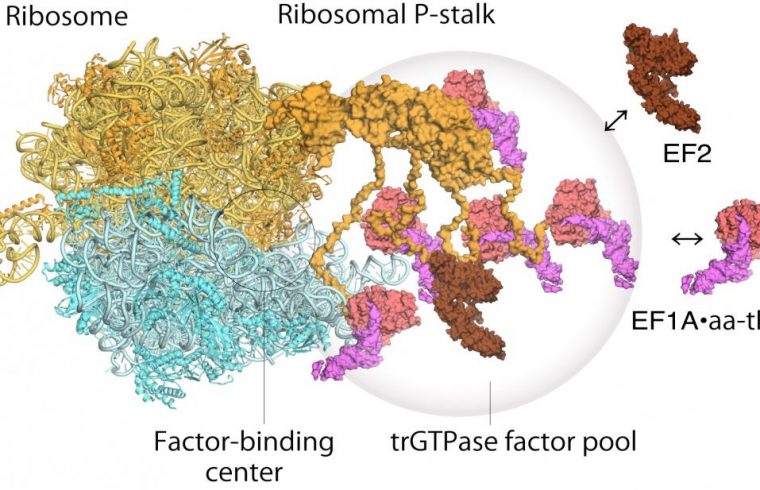
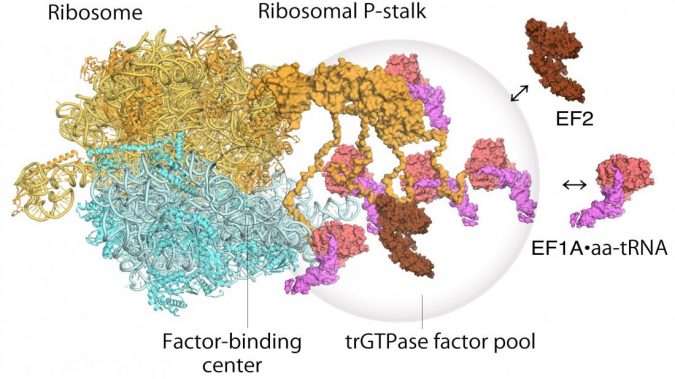
The ribosome stalk protein in particular plays an integral role in the protein synthesis process by recruiting protein factors responsible for translation and elongation of the amino acid sequence.
However it has been hard to satisfactorily establish the structure of the bound ribosome stalk protein because of its flexibility. Here the high resolution and fast image capture of high-speed atomic force microscopy proved invaluable.
Atomic force microscopy uses a nanoscale tip to feel samples, much like a vinyl record player needle scanning over a record, except that the details identified by an atomic force microscope can have atomic-scale resolution.
The versatility of the technique for different surfaces was already a huge advantage for biological studies, but with the advent of high-speed atomic force microscopy the technique was able to capture dynamic processes for the first time as well. Imai, Uchiumi and Kodera used the technique to reveal that the stalk protein actually flips between two conformations – one that agrees with previous structural models and one entirely unexpected new conformation.
As for how the ribosome operates, a two step mechanism had been previously proposed to describe how genetic information is translated through proteins known as “translational GTPase factors”.
The first step is the recruitment of the factors to the factor-tethering site on the protein stalk, thereby increasing the concentration of factors there – so-called factor pooling.
The second step is the binding and stabilizing of a translational GTPase on the ribosomal factor-binding center to catalyse GTPase hydrolysis.
From their high speed atomic force microscopy study the researchers were able to obtain the first visual evidence for the translational GTPase factor pooling mechanism by the ribosomal stalk.
Although the study was unable to give conclusive evidence of the action of the factors once bound, the researchers did note that the factors appeared to be retained in the vicinity once GTPase hydrolysis was complete, suggesting a potential role of the stalk protein in further stages of protein synthesis.
The researchers conclude, “future work with HS-AFM will provide further important information to understand the dynamic behaviors of these complex translational machineries.”