 Scientists from the Scripps Research Institute created an efficient screening process that can quickly find new “enediyne natural products,” which are essentially promising drug leads derived from soil microbes.
Scientists from the Scripps Research Institute created an efficient screening process that can quickly find new “enediyne natural products,” which are essentially promising drug leads derived from soil microbes.
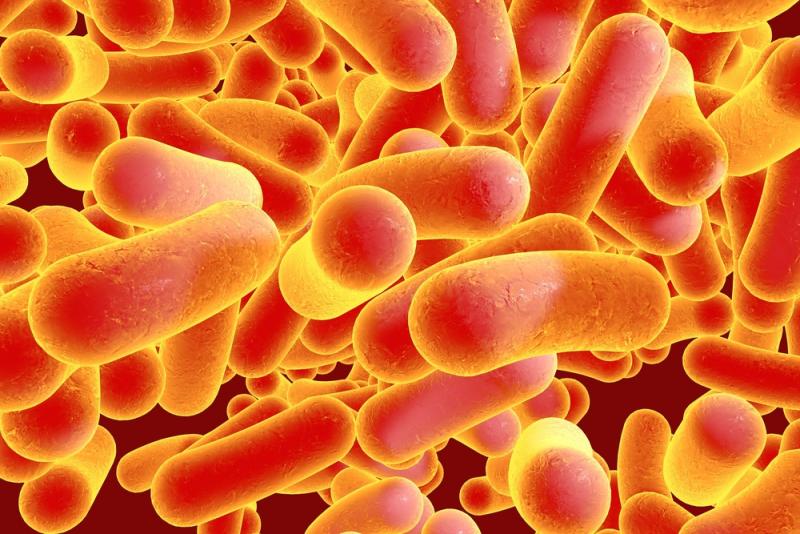
The team, led by TSRI professor Ben Shen, Ph.D., used this technique to identify a new group of enediyne natural products named tiancimycins. These products can kill certain cancer cells more quickly and completely than the monoclonal antibodies used in conjunction with cytotoxic drugs that target only cancer cells, according to the official announcement.
“The enediynes represent one of the most fascinating families of natural products for their extraordinary biological activities,” said Shen in a statement. “By surveying 3,400 strains from the TSRI collection, we were able to identify 81 strains that harbor genes encoding enediynes. With what we know, we can predict novel structural insights that can be exploited to radically accelerate enediyne-based drug discovery and development.”
Essentially, Shen’s method involved prioritizing the microbial strains that could produce the most important natural compounds enabling a more efficient allocation of resources during the discovery process.
Other researchers have used similar methods to sift through large amounts of data in order to pinpoint promising compounds to treat diseases.
A team at the Broad Institute of MIT and Harvard performed a similar process where they searched through a large chemical compound database for experimental malaria drugs with previously unknown novel mechanisms of action.
Shen and his colleagues published their work in the journal mBIO.
Source: dddmag.com